Diagnostics
Diagnostic assays for marine mammal parasites can be used to diagnose animals in rehabilitation. Marine mammals, when sick, often become stranded on beaches and are taken in by rehabilitation facilities. Once diagnosed, they can be provided with specialized treatment plans and hopefully be released back into the wild once they have recovered. We design molecular diagnostic assays that are specific, sensitive, and as non-invasive to the animal as possible. Many of our assays rely on feces or sputum (although some require blood), which would make them ideal candidates for surveying parasite infection levels in wild populations.
Sea lion Lungworm
A qPCR assay for Parafilaroides decorus, an otariid lungworm, using feces or sputum
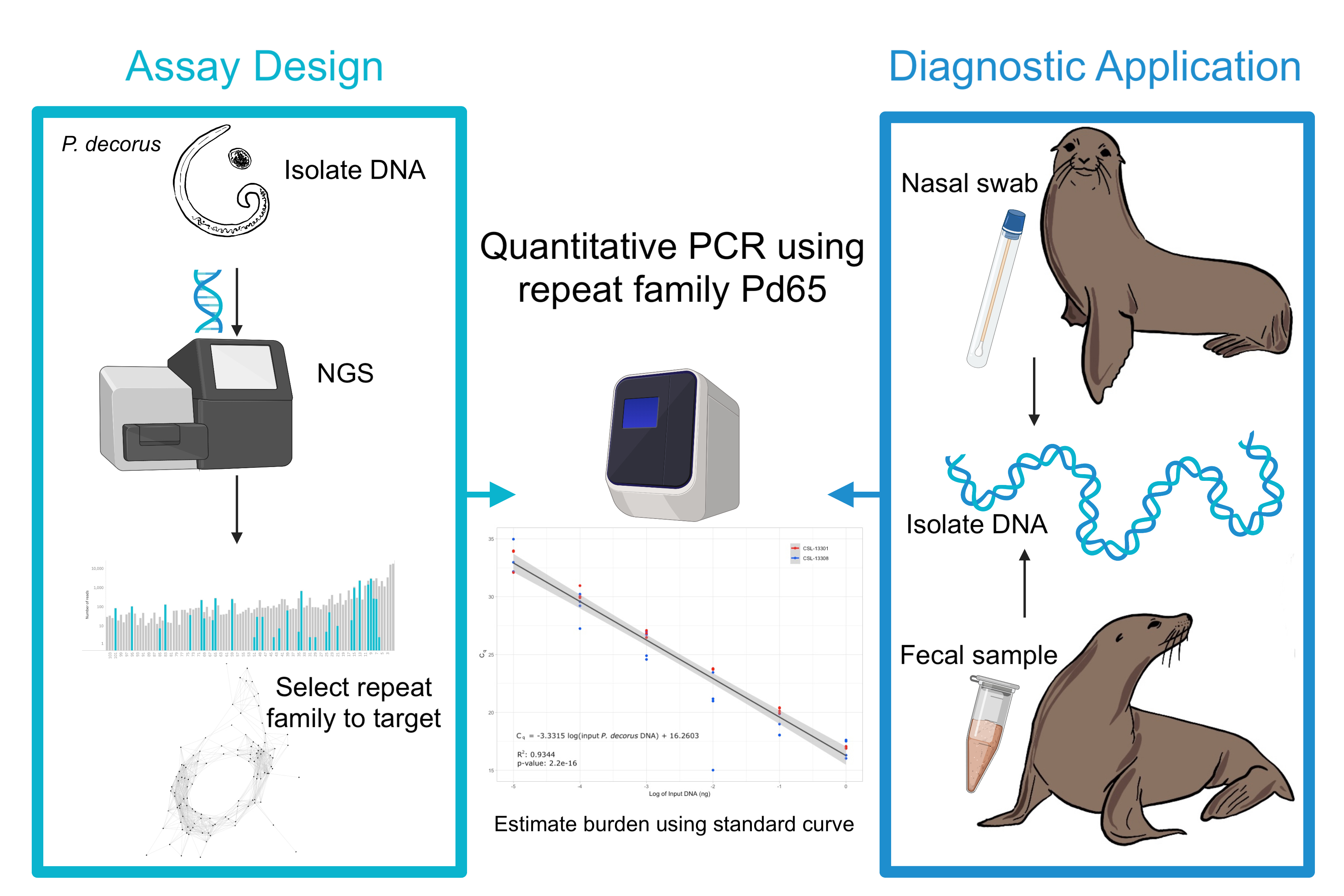
Abstract
Parafilaroides decorus, also known as sea lion lungworm, is a metastrongyloid nematode that infects otariid hosts, such as the charismatic California sea lion, Zalophus californianus. P. decorus causes bronchointerstitial pneumonia, respiratory distress, reduced ability to swim, dive and hunt and as a result, increased mortality particularly in young animals. Respiratory disease is a leading cause of stranding and admission to rehabilitation centers on the Pacific coast. Low-coverage genomic sequencing of four P. decorus individuals analyzed through Galaxy’s RepeatExplorer identified a novel repeat DNA family we employed to design a sensitive quantitative PCR (qPCR) assay for diagnosing infections from fecal or sputum samples. The assay detects as little as 10 fg of P. decorus DNA and a linear regression model developed using a standard curve can be used to estimate the concentration of P. decorus DNA in a sample, ± 0.015 ng. This knowledge can be leveraged to estimate the level of parasite burden, which can be used to design improved treatments for animals in rehabilitation. Improved treatment of infections will aid in more animals being successfully released back into the wild.
Seal Heartworm
A qPCR assay for seal heartworm, Acanthocheilonema spirocauda, using blood

Abstract
The distinct evolutionary pressures faced by Pinnipeds have likely resulted in strong coevolutionary ties to their parasites (Leidenberger et al., 2007). This study focuses on the phocid seal filarial heartworm species Acanthocheilonema spirocauda. A. spirocauda is known to infect a variety of phocid seals, but does not appear to be restricted to a single host species (Measures et al., 1997; Leidenberger et al., 2007; Lehnert et al., 2015). However, to date, seal heartworm has never been reported in grey seals (Halichoerus grypus) (Measures et al., 1997; Leidenberger et al., 2007; Lehnert et al., 2015). The proposed vector for seal heartworm is Echinophthirius horridus, the seal louse. Seal lice are known to parasitize a wide array of phocid seal species, including the grey seal. With the advent of climate change, disease burden is expected to increase across terrestrial and marine mammals (Harvell et al., 2002). Accordingly, increased prevalence of seal heartworm has recently been reported in harbor seals (Phoca vitulina) (Lehnert et al., 2015). Thus, the need for improved, rapid, and cost-effective diagnostics is urgent. Here we present the first A. spirocauda-specific rapid diagnostic test (a quantitative real-time PCR assay), based on a highly repetitive genomic DNA repeat identified using whole genome sequencing and subsequent bioinformatic analysis. The presence of an insect vector provides the opportunity to develop a multifunctional diagnostic tool that can be used not only to detect the parasite directly from blood or tissue specimens, but also as a molecular xenomonitoring (XM) tool that can be used to assess the epidemiological profile of the parasite by screening the arthropod vector. Using this assay, we provide evidence for the first reported case of seal heartworm in a grey seal.
Wolbachia: an Endosymbiont of Nematodes
Horizontal Gene Transfer (HGT)
Ancient HGT from Wolbachia in Seal Heartworm
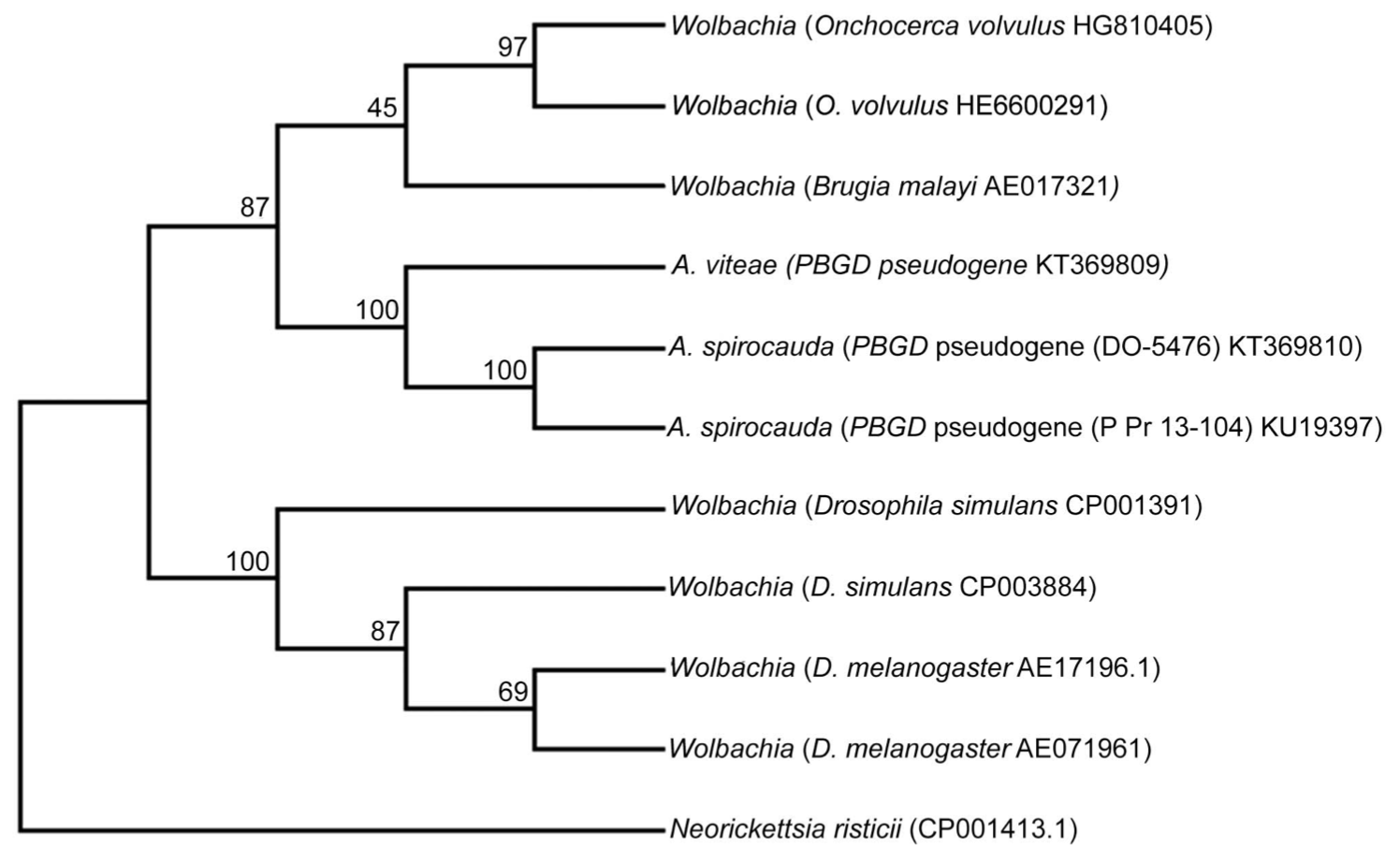
Abstract
The symbiotic relationship of Wolbachia spp. was first observed in insects and subsequently in many parasitic filarial nematodes. This bacterium is believed to provide metabolic and developmental assistance to filarial parasitic nematodes, although the exact nature of this relationship remains to be fully elucidated. While Wolbachia is present in most filarial nematodes in the family Onchocercidae, it is absent in several disparate species such as the human parasite Loa loa. All tested members of the genus Acanthocheilonema, such as Acanthocheilonema viteae, have been shown to lack Wolbachia. Consistent with this, we show that Wolbachia is absent from the seal heartworm (Acanthocheilonema spirocauda), but lateral gene transfer (LGT) of DNA sequences between Wolbachia and A. spirocauda has occurred, indicating a past evolutionary association. Seal heartworm is an important pathogen of phocid seals and understanding its basic biology is essential for conservation of the host. The findings presented here may allow for the development of future treatments or diagnostics for the disease and also aid in clarification of the complicated nematode–Wolbachia relationship.